Written 2016-10-30
The following was written as my (scathing) criticism of the Build-A-Cell initiative. I shared it Drew Endy and his lab, and actually ended up getting invited to work there, so I guess it ended up OK. I essentially argued that their current initiative would not foster innovation because of structural problems. As of 2021, right as I am writing this, I think that was a fair criticism.
Back then, they were looking at using E.coli mini-cells, and then building out a full cell from there. The key takeaway from me would be:
it makes more sense to use a top down approach because the end goals of growth are the consistent with the method of building. Bottom up, on the other hand, selects for specific functions not contextualized to growth.
Build-A-Cell is an project aimed at "Engineering an understandable cell." More information can be found on their website, buildacell.io. Conceptually, the idea is to create a modular 'biokernel', which is a core genome that is able to grow and divide, in which 'modules' are added to to accomplish more complex biosynthetic processes. For more information on their design, please follow the link above. Here, I, Keoni Gandall, discuss my opinions on the project's 'biokernel'.
Build-A-Cell website, Accessed 30 October 2016
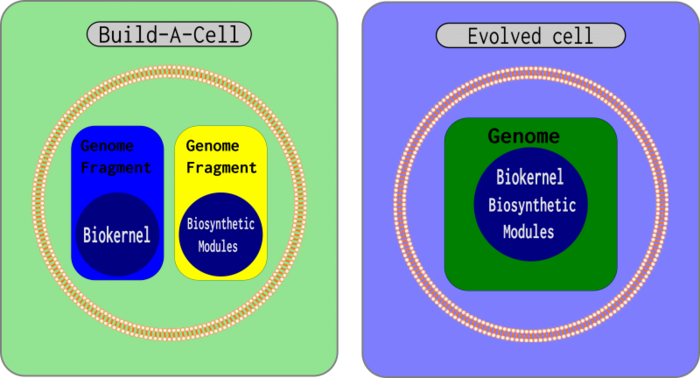
The aim is to create "a cell-like object containing native cellular machinery but lacking native DNA—using a rationally designed, forward engineered genome to restore growth and division." However, it is arguable that the objective, other than rational design, has been accomplished already with genome transplantation, by a top down removal of genes. However, the goal is best stated as a "rationally designed, forward engineered genome". Their "rationally designed" cell lacks a crucial component: an understood 'biokernel'. A well understood 'biokernel' would likely be a pragmatically minimal genome, optimized for growth and modularity. A rationally designed genome should not have any gaps in our understanding of its function, and therefore, as well focus on 'module development', there should be focus on a 'biokernel', as the 2 goals of 'modules' and of 'biokernel' are incompatible in their discovery.
In computers, there are 2 different forms of kernels: A monolithic kernel and a microkernel. Briefly, monolithic kernels run as a single program, increasing efficiency, while microkernels run in separate user spaces, increasing modularity and crash resistance. It is important to note that these architectures are inherently NOT biological, but human-made for the purpose of modelability and efficiency. A biological system cannot effectively replicate this structure, since they are 2 fundamentally different systems.
In order to have a biological system that has all functions known, a pragmatic minimal cell would be desired, upon which biological modules could be built onto. A pragmatic minimal cell would be one that "forms the core of a genetic operating system in which we understand the function and regulation of all genes", while also growing rather quickly, in order to facilitate efficiency in all downstream applications.
There are 2 facts that should be considered:
Minimal viable products are fundamental in Startup business models, and can cut the cost of failed ideas or projects. If a minimal viable product can be achieved, the feasibility of Build-A-Cell is realized, instead of just conjectured about, which would assist in convincing academics and industrial partners to adopt the Build-A-Cell system.
A biokernel is defined as the minimal set of genes necessary for growth.
If the stated goal is to create biokernel, it makes more sense to use a top down approach because the end goals of growth are the same. A bottom up approach selects for non contextualized function NOT growth, and thus can be flawed in it's testing. If flawed, which I will elaborate on in "Venter's Lessons of Designability", the project may reach an impenetrable barrier to a minimal viable product.
Given that a minimal viable product is essential for academic and industrial partnerships, Build-A-Cell may be doomed to failure if one cannot be achieved. The following are more reasons why a bottom up approach could be inferior to a top down approach.
The Build-A-Cell project proposes to use "Bottom Up" and "Top Down" approaches, "combin[ing] the best parts of both approaches". This involves a workflow to test genes in both a genome and genome-less environment.
In CRISPRi-ed E coli strains, desired genes are knocked out with CRISPR and then replaced with a synthetic gene as complementation. Although this is a simple process, it does not necessarily eliminate the original gene from the organism of question. There could be leakiness due to unknown transcriptional start and stop sites. In order to get a truly complete system, genes would have to be completely replaced, which is a non-trivial task for a non-modularized and large genome, such as E coli's. A mutation in an unknown gene promoting the gene expression of a module is far more likely if the native module is still in the genome than if it is deleted.
In addition, the majority of essential genes are in operons. CRISPRi knockdowns affect all downstream transcription, and therefore it is an unclean system for deleting and replacing genes.
These tests are still useful, however, because are contextualized to cell growth. A genome in a minicell, on the other hand, is completely isolated from the context of an actual cell, and this can lead to problems later in the project when a bottom up design is desired.
The Venter group has attempted in the past to use a "bottom up" approach to design their minimal genome. At this time (2016), "Preliminary knowledge-based HMG [Hypothetical Minimal Genome] design does not yield a viable cell". Overall, until there is full understanding of a minimal organism, rational design of one results in a non-functional genome. The Venter group also made part of the genome modular, a stated goal of the Build-A-Cell project. In order to get their minimal and partially-modular genome, the group had to run several parts of the genome in native contexts to be able to isolate variables that caused cell death. This, by definition, would be a "top down" approach, yet it is the only one that so far has worked to get a core set of genes. In order to get a successful 'biokernel' a similar process should be taken to define a 'core' of the genetic operating system.
Design and synthesis of a minimal bacterial genome (2016)
A "bottom up" approach, using testing in genome-less minicells, is essentially that of the Hypothetical Minimal Genome that Venter's group intended to make, which did not work. Given that E coli is an even more complex organism than the Mycoplasmas that Venter worked with, it seems unlikely that a successful boot process will be able to take place without first verifying what genes are required for said boot process using a "top down" process. In finding what genes are completely necessary to boot, one would create a 'biokernel'.
For example, the MiniBacillus project aims to create a minimal Bacillus cell through serial integrations of DNA. In order to get Bacillus subtilis to the size of a basic Mycoplasma, under 1mb, it would require approximately 52 integrations. This genome would be created by integration of cassettes, not through true 'genome replacement'. This is an unwieldy amount of integrations to do on a genome when there are viable alternatives. A question to answer is - will it be easier to create basic tools for a different organism of 1/5 the size, or use a studied organism? Given that large scale rearrangement and individual testing is still required, the first is more likely to be a viable project in a shorter amount of time.
Once a genome with core genes is created and identified, modularization can take place. The ability to clone a large amount of genomes quickly is paramount. In order to do this, a genome would hypothetically not require any large scale synthesis or PCR once modules are created. This includes CRISPRi cloning and verification of proper repression, which are 2 things which do not lend themselves easily to brute modularization. By creating a large amount of genomes, the modules could be systematically tested for 'modelability' without going through CRISPRi troubleshooting, and large scale reorganization of the genome could take place, directly leading to "rigorously characterised drivers of core functions and regulators of resources". In this way, modularization is the key to the stated goals of "Modularity, modelability, and simplicity of understanding".
My writings on the Modular genome
Modularization is what Build-A-Cell is looking for in a genome because it would accomplish all of the stated goals of the project. Modularization quickens testing of bottom up genomes while still keeping the advantages of top down positive controls. A modularized genome, with modules being genes, could also be distributed to a large range of labs for testing in a way module+CRISPRi technology can't be. Reproduciblity is an increasing problem in life sciences, and because of the inherent leakiness of using minicells and CRISPRi, there could very well be many holes in our understanding due to normal lab variation on results. Since a modular genome has only genetic information to be transplanted, context and leakiness of systems are less of a concern, since large scale verification trials can go on using a basis of growth media, instead of specialized module-specific conditions. This helps in reproducibility, which is essential in an open source project.
The choice of using an E coli cell as a minicell is debatable. While it has the best characterized genetic systems, all genomes would be cloned in it before transplantation. The synthetic genomes, therefore, unless isolated by clever use of orthogonal RBSs or transcriptional factors, would be nonorthogonal to the organism they are cloned in. Since orthogonality of transcription or translational start would be difficult to achieve, reducing modelability, and nonorthogonality leads to less modelability and simplicity, a logical choice would be to use an organism that is orthogonal to the organism that will be used to clone the genomes. For this reason, and a few others, the Venter group uses Yeast in the final cloning steps of their synthetic genome. A translationally isolated genome can potentially be one day created using the 57 codon E coli, but this project is still being worked on, and it does not seem like the most simple answer. Mycoplasmas, and the related Mesoplasmas, however, are already translationally isolated from E coli due to tryptophan being coded with a TGA codon. By using a Mollicute, an orthogonal genome can be created in E coli, the host which would be used for cloning and conjugation.
Cloning whole bacterial genomes in yeast
Usage of Minicells as containers for genomes is clever, however it hearkens the question of where exactly testable genomes should come from. Because E coli is the original host, it would be debatable if containers are even required, as genes and modules can be tested in the genome itself in a CAGE-like fashion. Testing of modules in a genome is a more direct way to elucidate precise biological mechanics of a module than a decontextualized minicell or a CRISPRi screen is.
Precise manipulation of bacterial chromosomes by conjugative assembly genome engineering
Build-A-Cell aims to create a biokernel with great modelability and simplicity of understanding. However, by using E coli, a large organism, these 2 values are compromised. In essence, simplicity of understanding and modelability are synonyms for the same idea - the idea of being able to create a cell from the bottom up. History has shown that a top down approach is needed to achieve minimal bottom up assembly, and thus I question the usefulness of stressing a bottom up approach, since a top down approach is needed to actually characterize what is required from the bottom up.
Because the proposed synthetic genome would be E coli, I question the usefulness of characterizing containers. Since cloning will be done in E coli, if a whole genome is used, a container will not be necessary. If the genome is on a BAC, it would be simpler to delete the native genome inside of that cell instead of moving it to a different chassis that you cannot effectively set up a cellular context in.
As an alternative direction for the Build-A-Cell, I would encourage an effort of minimalization and modularization of E coli, in which different teams could be assigned to different portions of the genome with competition to see who could modularize and minimalize their given 20-100kb the best. The information gathered from such a competition could be combined with CAGE to more effectively create a 'biokernel'. Each year, before the competition, an E coli strain with last year's results would be constructed for the teams of the current year to work with. In comparison to the current workflow of Build-CRISPRi-Test, this method would foster a more competitive spirit, and ultimately would create a more useful genome than simply testing modules would.
On a more personal direction, I would encourage adoption of Mesoplasma florum, a BSL1 organism, or a related species for Build-A-Cell. It is five times smaller than E coli, meaning it is simpler to understand and easier to modularize. Modularization of a genome would solve the stated goals of "modularity, modelability, and simplicity of understanding"[2] and since Mesoplasma is both orthogonal and closer to a 'biokernel' than E coli is, it could very well be a viable alternative as a 'biokernel'. As parts of E coli are deleted, undeniably the growth rate will decrease. Mesoplasma florum grows quickly at it's already reduced genome size of 800kb, with a doubling time of 40 minutes, possibly in comparatively simple media. Using this organism would put the Build-A-Cell closer to it's stated goals than many man-years of work with E coli will. By investing those man-years into full-genome modularization and distribution of Mesoplasma, a 'biokernel' as desired could be completed years quicker than one would be for E coli. For this reason, I encourage usage of Mesoplasma florum as an alternative for a biokernel.